Initial Publication Date: May 9, 2006
Methods employed at the Microbial Observatory:
The Oceanic Microbial Observatory combines oceanographic microbial ecological methodology with molecular microbiological /ecological expertise to elucidate the linkages between phylogenetic diversity and large scale biogeochemical processes.
Research Techniques:
Tangential Flow Filtering (TFF): In using TFF large volumes of sample can be concentrated by passing them over a filter of specific size. Particulates of the size desired are retained in a loop system by exerting pressure to force excess liquid, along with particles smaller than those of interest, through the tangential flow filter and out of the system. In a circular fashion the retained sample loses volume until the particles (in this case Bacteria or virus) of interest are concentrated enough to be worked with in the lab. Craig Carlson and Bob Morris are shown size fractionating dissolved organic carbon (DOC) via the TFF system aboard the R.V. Weatherbird II in June 2005.
Fluorescence In Situ Hybridization (FISH): FISH is a molecular technique that is used at the Oceanic MO to identify and enumerate specific bacterial groups. FISH works by designing a DNA or RNA probe to detect the presence of the complementary DNA sequence specific to target groups of interest. The complementary RNA, in this case, is in a variable portion of the 16S ribosomal RNA which is present only in a distinct bacterial group. See SAR11 clade dominates ocean surface bacterioplankton communities (Nature 420:806-810, 2002) for the use of FISH in enumerating SAR11 populations.
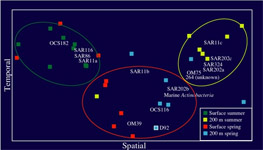
Terminal Restriction Fragment Length Polymorphism (T-RFLP): Along with bulk nucleic acid hybridization analyses T-RFLP is used to identify, characterize, and quantify spatial and temporal patterns in marine bacterioplankton communities at the study site. Terminal-Restriction Fragment Length Polymorphism analysis is a method of comparative community analysis. T-RFLP analysis is based on the restriction endonuclease digestion of fluorescently end-labeled PCR products (in this study the 16S rRNA gene). The digested products are separated by gel electrophoresis and detected on an automated sequence analyzer. The method provides distinct profiles (fingerprints) dependent on the species composition of the communities of the samples. Figure on left shows 'Non-metric Multidimensional Scaling' (NMS) of relative bacterial 16S rDNA terminal restriction fragments from monthly time-series samples collected at BATS between Feb and Sep (1992 and 2000).